Tools and techniques for managing running pipelines
eHive has an internal accounting system to help it efficiently allocate Workers and schedule Jobs. In normal operation, the Beekeeper updates the status of eHive components and internal statistics on an intermittent basis, without the need for intervention from the user. However, in some cases this accounting will not update correctly - usually when the user circumvents eHive’s automated operation. There are some utility functions available via the Beekeeper to correct these accounting discrepancies.
Synchronizing (“sync”-ing) the pipeline database
There are a number of Job counters maintained by the eHive system to help it manage the Worker population, monitor progress, and correctly block and unblock Analyses. These counters are updated periodically, in a process known as synchronization (or “sync”).
The sync process can be computationally expensive in large pipelines, so syncs are only performed when needed.
Rarely, the Job counters will become incorrect during abnormal pipeline
operation, for example if a few process crash, or after Jobs are manually
stopped and re-run. In this case, the user may want to manually re-sync the
database. This is done by running beekeeper.pl with the -sync option:
beekeeper.pl -url sqlite:///my_pipeline_database -sync
Re-balancing semaphores
When a group of Jobs is organized into a fan and funnel structure, the eHive
system keeps track of how many fan Jobs are not [ DONE ] or
[ PASSED_ON ]. When that count reaches zero, the semaphore controlling the
funnel Job is released. In some abnormal circumstances, for example when Workers
are killed without an opportunity to exit cleanly, eHive’s internal count of
Jobs remaining to do before releasing the semaphore may not represent reality.
Resetting Jobs in a fan is another scenario that can cause the semaphore count
to become incorrect. It is possible to force eHive to re-count fan Jobs and
update semaphores by running beekeeper.pl with the -balance_semaphores option:
beekeeper.pl -url sqlite:///my_pipeline_database -balance_semaphores
Garbage collection of dead Workers
On occasion, Worker processes will end without having an opportunity to update
their status in the eHive database. The Beekeeper will attempt to find these
Workers and update their status itself. It does this by reconciling the list of
Worker statuses in the eHive database with information on Workers gleaned from
the meadow’s process tables (e.g. ps, bacct, bjobs). This
process is called “garbage collection”. A typical output is like this:
Beekeeper : loop #12 ======================================================
GarbageCollector: Checking for lost Workers...
GarbageCollector: [Queen:] out of 66 Workers that haven't checked in during the last 5 seconds...
GarbageCollector: [LSF/EBI Meadow:] RUN:66
In this case, 66 LSF workers had not updated their status within the last 5
seconds but they were in fact all running and listed by bjobs. The
Garbage collection process ends there, then.
In another case, Beekeeper could find so called LOST Workers:
Beekeeper : loop #15 ======================================================
GarbageCollector: Checking for lost Workers...
GarbageCollector: [Queen:] out of 45 Workers that haven't checked in during the last 5 seconds...
GarbageCollector: [LSF/EBI Meadow:] LOST:4, RUN:41
GarbageCollector: Discovered 4 lost LSF Workers
LSF::parse_report_source_line( "bacct -l '4126850[15]' '4126850[6]' '4126835[24]' '4126850[33]'" )
GarbageCollector: Found why 4 of LSF Workers died
In this case, bjobs only listed 41 of the 45 LSF workers that had not
updated their status within the last 5 seconds. Beekeeper then had to
resort to bacct to find out what happened to 4 LOST Workers.
LOST Workers are most of the time Workers that have been killed by LSF
due to exceeding their allocated resources (MEMLIMIT or RUNLIMIT).
When no reason could be found, the cause of death recorded in the
log_message table will be UNKNOWN. This is known to happen when
bacct was executed too long after the Worker exited: LSF’s journal only
knows about the most recent jobs. It seems to be happening in other
circumstances that are not clearly understood. If you face this UNKNOWN
error, re-run the job locally in debug mode.
The Garbage collection happens at every Beekeeper loop, but a manual
reconciliation and update of Worker statuses can be invoked by running
beekeeper.pl with the -dead option:
beekeeper.pl -url sqlite:///my_pipeline_database -dead
Tips for performance tuning and load management
Resource optimisation
eHive automaticaly gathers resource usage information about every
workers and stores that in the worker_resource_usage table. Although it
can be accessed directly, the resource_usage_stats view gives a nicer
summary for each analysis:
> SELECT * FROM resource_usage_stats;
+---------------------------+-------------+--------------------------+-------------+---------+--------------+--------------+--------------+---------------+---------------+---------------+
| analysis | meadow_type | resource_class | exit_status | workers | min_mem_megs | avg_mem_megs | max_mem_megs | min_swap_megs | avg_swap_megs | max_swap_megs |
+---------------------------+-------------+--------------------------+-------------+---------+--------------+--------------+--------------+---------------+---------------+---------------+
| create_tracking_tables(1) | LSF | default(1) | done | 1 | 56.7109 | 56.71 | 56.7109 | NULL | NULL | NULL |
| MLSSJobFactory(2) | LSF | default_with_registry(5) | done | 1 | 67.5625 | 67.56 | 67.5625 | NULL | NULL | NULL |
| count_blocks(3) | LSF | default_with_registry(5) | done | 1 | 56.9492 | 56.95 | 56.9492 | NULL | NULL | NULL |
| initJobs(4) | LSF | default_with_registry(5) | done | 1 | 66.6445 | 66.64 | 66.6445 | NULL | NULL | NULL |
| createChrJobs(5) | LSF | default_with_registry(5) | done | 1 | 61.2188 | 61.22 | 61.2188 | NULL | NULL | NULL |
| createSuperJobs(6) | LSF | default_with_registry(5) | done | 2 | 61.1992 | 61.20 | 61.207 | NULL | NULL | NULL |
| createOtherJobs(7) | LSF | crowd_with_registry(4) | done | 1 | 100.398 | 100.40 | 100.398 | NULL | NULL | NULL |
| dumpMultiAlign(8) | LSF | crowd(2) | done | 52 | 108.848 | 695.60 | 1330.91 | NULL | NULL | NULL |
| emf2maf(9) | LSF | crowd(2) | done | 46 | 58.3008 | 132.88 | 150.695 | NULL | NULL | NULL |
| compress(10) | LSF | default(1) | done | 17 | 58.1758 | 58.24 | 58.2695 | NULL | NULL | NULL |
| md5sum(11) | LSF | default(1) | done | 2 | 58.2227 | 58.23 | 58.2344 | NULL | NULL | NULL |
| move_maf_files(12) | LSF | default(1) | done | 1 | 58.1914 | 58.19 | 58.1914 | NULL | NULL | NULL |
| readme(13) | LSF | default_with_registry(5) | done | 2 | 77.5859 | 83.13 | 88.668 | NULL | NULL | NULL |
| targz(14) | NULL | default(1) | NULL | 1 | NULL | NULL | NULL | NULL | NULL | NULL |
+---------------------------+-------------+--------------------------+-------------+---------+--------------+--------------+--------------+---------------+---------------+---------------+
In this example you can see how much memory each analysis used and decide how much to allocate them, e.g. 100 Mb for most analyses, 200 Mb for “createOtherJobs” and “emf2maf”, and 1,500 Mb for “dumpMultiAlign”. However, it seems that the average memory usage of “dumpMultiAlign” is below 700 Mb, meaning that more than half of the requested memory could be wasted each time. You can get the actual breakdown with this query:
> SELECT 100*CEIL(mem_megs/100) AS mem_megs, COUNT(*) FROM worker_resource_usage JOIN role USING (worker_id) WHERE analysis_id = 8 GROUP BY CEIL(mem_megs/100);
+----------+----------+
| mem_megs | COUNT(*) |
+----------+----------+
| 200 | 1 |
| 300 | 4 |
| 400 | 6 |
| 500 | 10 |
| 600 | 9 |
| 700 | 8 |
| 800 | 5 |
| 900 | 4 |
| 1000 | 2 |
| 1200 | 2 |
| 1400 | 1 |
+----------+----------+
You can see that about three quarters of the jobs used less than 700Mb, so another strategy is to give 700Mb to the analysis, expect some jobs to fail (i.e. to be killed by the compute farm) and wire a copy of the analysis with more memory on the -1 branch (MEMLIMIT), cf Special Dataflow when Jobs Exceed Resource Limits. You can chain with MEMLIMIT as many analyses as required to provide the appropriate memory usage steps, e.g.
{ -logic_name => 'Alpha',
-flow_into => {
-1 => [ 'Alpha_moremem' ],
},
},
{ -logic_name => 'Alpha_moremem',
-flow_into => {
-1 => [ 'Alpha_himem' ],
},
},
{ -logic_name => 'Alpha_himem',
-flow_into => {
-1 => [ 'Alpha_hugemem' ],
},
},
{ -logic_name => 'Alpha_hugemem',
},
|
![digraph test {
ratio="compress"; concentrate = "true"; name = "AnalysisWorkflow"; pad = "0";
analysis_Alpha [fillcolor="white", fontname="Times-Roman", label=<<table border="0" cellborder="0" cellspacing="0" cellpadding="1"><tr><td colspan="1">Alpha</td></tr></table>>, shape="Mrecord", style="filled"];
analysis_Alpha_himem [fillcolor="white", fontname="Times-Roman", label=<<table border="0" cellborder="0" cellspacing="0" cellpadding="1"><tr><td colspan="1">Alpha_himem</td></tr></table>>, shape="Mrecord", style="filled"];
analysis_Alpha_hugemem [fillcolor="white", fontname="Times-Roman", label=<<table border="0" cellborder="0" cellspacing="0" cellpadding="1"><tr><td colspan="1">Alpha_hugemem</td></tr></table>>, shape="Mrecord", style="filled"];
analysis_Alpha_moremem [fillcolor="white", fontname="Times-Roman", label=<<table border="0" cellborder="0" cellspacing="0" cellpadding="1"><tr><td colspan="1">Alpha_moremem</td></tr></table>>, shape="Mrecord", style="filled"];
analysis_Alpha -> analysis_Alpha_moremem [color="blue", fontcolor="blue", fontname="Helvetica", label="#-1\n"];
analysis_Alpha_himem -> analysis_Alpha_hugemem [color="blue", fontcolor="blue", fontname="Helvetica", label="#-1\n"];
analysis_Alpha_moremem -> analysis_Alpha_himem [color="blue", fontcolor="blue", fontname="Helvetica", label="#-1\n"];
subgraph "cluster_tmpl5m9wtpx" {
label="";
style="filled";
colorscheme="blues9";
fillcolor="1";
color="1";
analysis_Alpha;
analysis_Alpha_himem;
analysis_Alpha_hugemem;
analysis_Alpha_moremem;
}
}](../_images/graphviz-bea57e9a0b20d6025b818411a5e69d777fbba6d8.png) |
Relying on MEMLIMIT can be inconvenient at times:
The mechanism may not be available on all job schedulers (of the ones eHive support, only LSF has that functionality).
When LSF kills the jobs, the open file handles and database connections are interrupted, potentially leading in corrupted data, and temporary files hanging around.
Since the processes are killed in a random order and not atomically, sometimes, the child process (e.g. an external program your Runnable is running) will be killed first, and the Runnable will have enough time to record this job attempt as failed (but not as MEMLIMIT), take another job and then be killed, making eHive think it is the second job that has exceeded the memory requirement. On LSF we advice waiting 30 seconds when detecting that an external command has been killed to give LSF enough time to kill the worker too.
This is time-expensive since a job may be tried with several memory requirements before finally finding the right one.
Instead of relying on MEMLIMIT, a more efficient approach is to predict the
amount of memory the job is going to need. You would first need to
understand what is causing the high memory usage, and try to correlate that to
some input parameters (for instance, the length of the chromosome, the
number of variants, etc). Then you can define several resource classes and
add WHEN conditions to the seeding dataflow to wire each job to the right
resource class.
Here is an example from an Ensembl Compara pipeline:
-flow_into => {
"2->A" => WHEN (
"(#total_residues_count# <= 3000000) || (#dnafrag_count# <= 10)" => "pecan",
"(#total_residues_count# > 3000000) && (#total_residues_count# <= 30000000) && (#dnafrag_count# > 10) && (#dnafrag_count# <= 25)" => "pecan_mem1",
"(#total_residues_count# > 30000000) && (#total_residues_count# <= 60000000) && (#dnafrag_count# > 10) && (#dnafrag_count# <= 25)" => "pecan_mem2",
"(#total_residues_count# > 3000000) && (#total_residues_count# <= 60000000) && (#dnafrag_count# > 25)" => "pecan_mem2",
"(#total_residues_count# > 60000000) && (#dnafrag_count# > 10)" => "pecan_mem3",
),
"A->1" => [ "update_max_alignment_length" ],
},
Resource usage overview
The data can also be retrieved with the generate_timeline.pl script in the form of a graphical representation:
generate_timeline.pl --url $EHIVE_URL --mode memory -output timeline_memory_usage.png
generate_timeline.pl --url $EHIVE_URL --mode cores -output timeline_cpu_usage.png
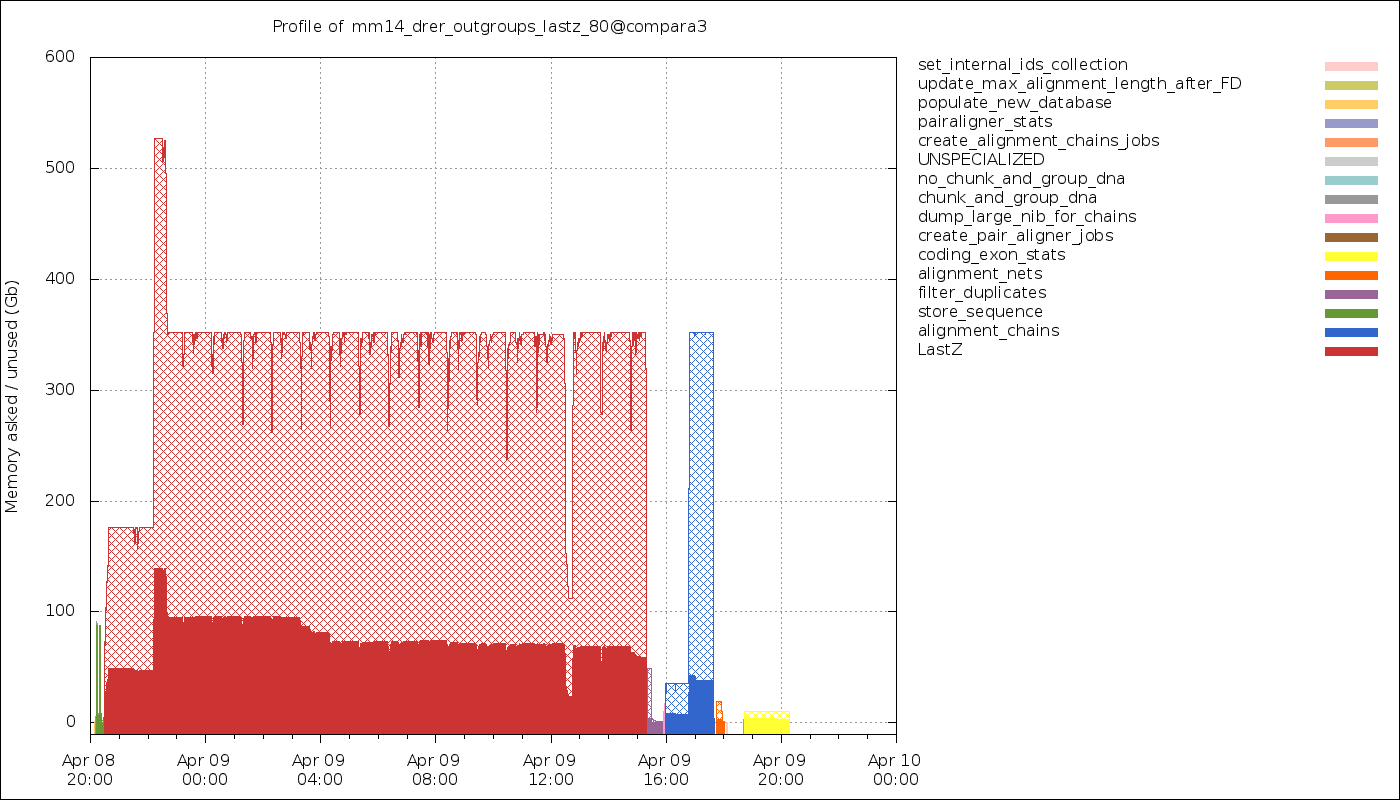
Timeline of the memory usage. The hatched areas represent the amount of memory that has been requested but not used.
Since eHive forces you to bin jobs into a smaller number of analyses, each analysis having a single resource class (a memory requirement), each job may not run with the exact amount of memory it needs. Some level of memory over-reservation is expected (although the plot above shows too much of that !).
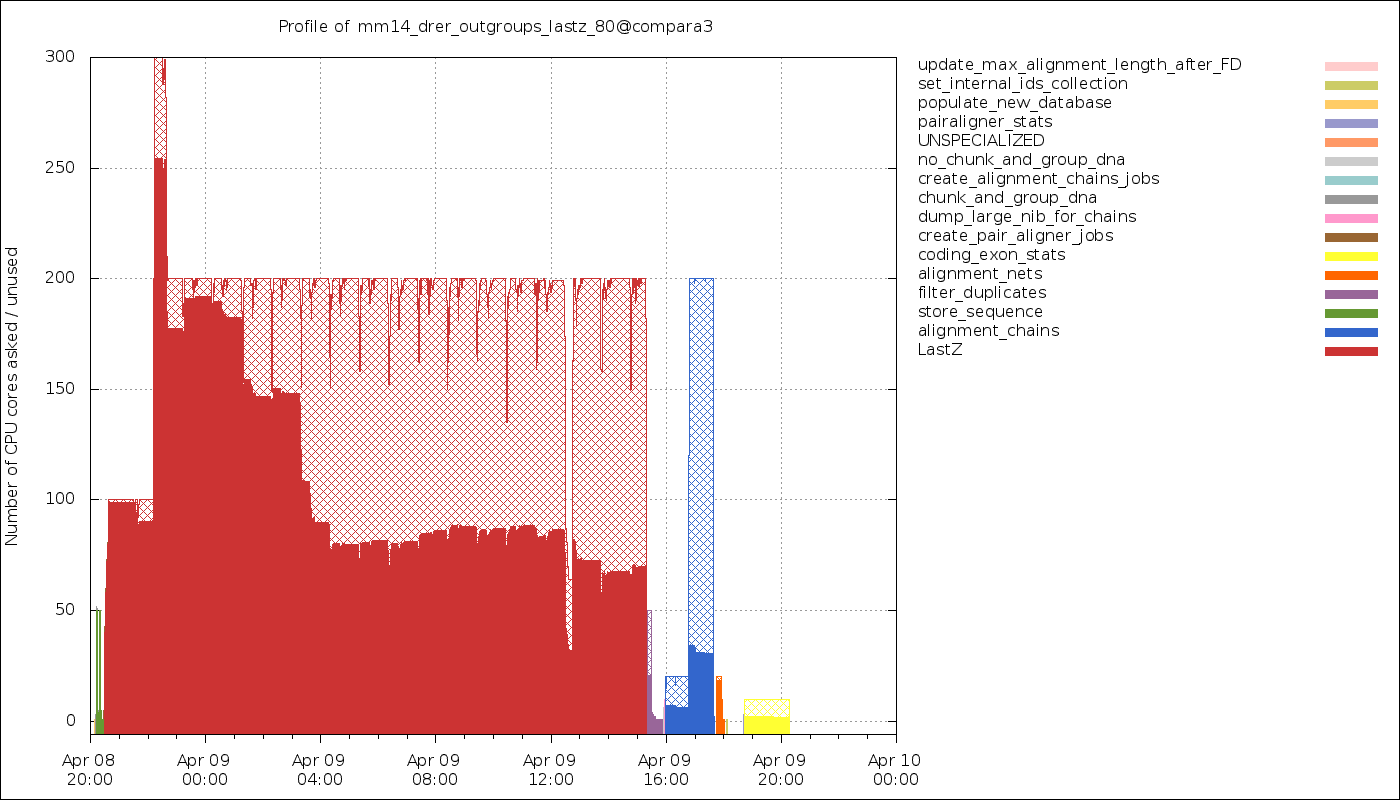
Timeline of the CPU usage. The hatched areas represent the fraction of the wall time spent on sleeping or waiting (for I/O, for instance).
It is most of the time expected to not be fully using the CPUs, as most jobs will have to read some input data and write some results. both of which subject to I/O waits. You also need to consider that all the SQL queries you will be sending to a database server (either directly or via an Ensembl API) will shift the focus to the server and make your own Runnable wait for the result. Finally, many job schedulers (such as LSF) can only allocate whole CPU cores, meaning that even if you estimate you only need 50% of a core, you might be forced to still allocate 1 core and “waste” the other 50%.
Capacity and batch size
A number of parameters can help increasing the performance of a pipeline, but capacities and batch sizes have the most direct effect. Both parameters go hand in hand.
Although Workers run Jobs one at a time, they can request (claim) more than one Job (a batch) from the database. It means a Worker would successively have:
Jobs claimed, 0 running, 0 done
-1 Jobs claimed, 1 running, 0 done
-1 Jobs claimed, 0 running, 1 done
-2 Jobs claimed, 1 running, 1 done
-2 Jobs claimed, 0 running, 2 done
etc.
It is useful as long as claiming 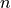 Jobs at a time is faster than
claiming
Jobs at a time is faster than
claiming 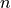 times 1 Job, and that the claiming process doesn’t lock
the table for too long (which would prevent other Workers from operating
normally).
times 1 Job, and that the claiming process doesn’t lock
the table for too long (which would prevent other Workers from operating
normally).
This can mitigate the overhead of submitting many small, fast-running Jobs to the farm. Bear in mind that increasing the batch size helps relieving the pressure on the Job table from claiming Jobs only. As the Job table is used to track the current status of jobs, it can also be slowed down by running too many Workers, regardless of the batch size. And more generally, the Jobs may create additional load on other databases, filesystems, etc, which are your responsibility to monitor.
Optimizing the batch size is something of an art, as the optimal size is a function of Job runtime and the number of Jobs in contention for the eHive database. Here follows some estimates of the optimal parameters to run a single Analysis, composed of 1 million Jobs, under two scenarios:
Best throughput: the combination of parameters that gets all the Jobs done the fastest.
Best efficiency: the combination of parameters that has the highest capacity whilst maintaining an overhead per Job below 10 milliseconds. The overhead is defined as the average amount of time eHive spends per Job for general housekeeping tasks, but also for claiming. For instance, a Worker that has lived 660 seconds and run 600 Jobs (each set to sleep 1 second) will have an overhead of 10 millisecond per Job. eHive has a minimum overhead per Job of 6-7 milliseconds.
In general, “best throughput” parameters put a lot more pressure on the database. Only use these parameters if you are in a rush to get your Analysis done, and if you are allowed to use that much resources from the server (the server might be unable to run someone else’s pipeline at the same time !).
Job’s duration |
Best efficiency |
Best throughput |
|||||
|---|---|---|---|---|---|---|---|
Capacity |
Batch size |
Analysis duration |
Capacity |
Batch size |
Job overhead |
Analysis duration |
|
50 ms |
25 |
20 to 1,000 |
2,315 s |
100 |
200 |
58 ms (116%) |
1,080 s |
100 ms |
50 |
20 to 1,000 |
2,185 s |
100 |
200 |
22 ms (22%) |
1,215 s |
500 ms |
100 |
10 to 500 |
5,085 s |
250 |
100 |
16 ms (3.2%) |
2,055 s |
1 s |
250 |
10 to 50 |
4,040 s |
500 |
50 |
257 ms (25.7%) |
2,515 s |
5 s |
500 |
20 |
10,020 s |
2,500 |
20 |
545 ms (10.9%) |
2,220 s |
These values have been determined with a pipeline entirely made of Dummy Jobs (they just sleep for a given amount of time) at various capacities (1, 5, 10, 25, 50, 100, 200 or 250, 500, 1,000, 2,500, 5,000) and batch sizes (1, 2, 5, 10, 20, 50, 100, 200, 500, 1,000, 2,000, 5,000), for various sleep times. The notion of sleep models operations on other databases (data processing with the Ensembl API, for instance), running a system command, etc. Although the actual results are specific to the MySQL server used for the benchmark, the trend is expected to be the same on other versions of MySQL.
Hive capacity vs Analysis capacity
Analysis capacity
Limits the number of Workers that
beekeeper.plwill run for this particular Analysis. It does not mean if you set it to 200 there will be exactly 200 workers of this Analysis, as there are other considerations taken into account by the scheduler, but there will be no more than 200.
hive capacity
Also limits the number of Workers, but globally across the whole pipeline. If you set -hive_capacity of an Analysis to X it will mean “one Worker of this Analysis consumes 1/X of the whole Hive’s capacity (which equals to 1.0)”. Like Analysis capacity, setting it to 200 means that you will not get more than 200 running Workers. Using it only makes sense if you need several Analyses running in parallel and consuming the same resource (e.g. accessing the same table) to balance load between themselves.
If one of these is set to 0, eHive will not schedule any Workers for the Analysis (regardless of the value of the other parameter). If a parameter is not set (undefined), then its related limiter is unused.
Examples
analysis_capacity=0 and hive_capacity is not set:
No Workers are allowed to run
analysis_capacity=0 and hive_capacity=150:
No Workers are allowed to run
analysis_capacity is not set and hive_capacity=0:
No Workers are allowed to run
analysis_capacity is not set and hive_capacity=150:
No Workers are allowed to run
analysis_capacity=150 and hive_capacity is not set:
eHive will schedule at most 150 Workers for this Analysis
A.hive_capacity=1 and B.hive_capacity=300. Examples of allowed numbers of Workers are:
Analysis A
Analysis B
1
0
0
300
A.hive_capacity=100, A.analysis_capacity=1 and B.hive_capacity=300. Examples of allowed numbers of Workers are:
Analysis A
Analysis B
1
297
0
300
A.hive_capacity=100 and B.hive_capacity=300. Examples of allowed numbers of Workers are:
Analysis A
Analysis B
100
0
75
75
50
150
25
225
0
300
A.hive_capacity=100, B.hive_capacity=300 and B.analysis_capacity=210. Examples of allowed numbers of Workers are:
Analysis A
Analysis B
100
0
75
75
50
150
30
210
More efficient looping
The Beekeeper is not constantly active: it works a bit, up to several seconds
depending on the size and complexity of the pipeline, and the
responsiveness of the job scheduler, and then sleeps for a given amount of
time (by default 1 minute, the -sleep parameter). Every loop, the Beekeeper
submits -submit_workers_max Workers (which defaults to 50), to avoid
overloading the scheduler with submitted Jobs.
You can change both parameters, for instance reduce the sleep time to submit
Workers more frequently (e.g. 12 seconds == 0.2 minutes), or increase
-submit_workers_max to submit more Workers every loop (e.g. 100 or
200) as long as the server supports it. It is good practice to give
Workers time to check-in with the eHive database between loops. The default
parameters are safe values that generally work well for production
pipelines.
The impact of loop time on the overall time to complete a workflow will be fairly small, however. When a Worker completes a Job, it looks for new Jobs that it can run, and will claim and run them automatically - the Beekeeper is not involved in this claiming process. It’s only in the case where new Workers need to be created that the pipeline would be waiting for another Beekeeper loop.
Other limiters
Besides the Analysis-level capacities, the number of running Workers is
limited by the TotalRunningWorkersMax parameter. This parameter has a
default value in the a hive_config.json file in the root of the eHive
directory and can be changed at the Beekeeper level with the --total_running_workers_max option.
Every time the Beekeeper loops, it will check the current state of your
eHive pipeline and the number of currently running Workers. If it
determines more Workers are needed, and the -total_running_workers_max
value hasn’t been reached, it will submit more, up to the limit of
-submit_workers_max.
Database server choice and configuration
SQLite can have issues when multiple processes are trying to access the database concurrently because each process acquires locks the whole database. As a result, it behaves poorly when the number of workers reaches a dozen or so.
MySQL is better at those scenarios and can handle hundreds of concurrent active connnections. In our experience, the most important parameters of the server are the amount of RAM available and the size of the InnoDB Buffer Pool.
We have only used PostgreSQL in small-scale tests. If you get the chance to run large pipelines on PostgreSQL, let us know ! We will be interested in hearing how eHive behaves.
Database connections
The Ensembl codebase does, by default, a very poor job at managing database connections, and how to solve the “MySQL server has gone away” error is a recurrent discussion thread. Even though eHive’s database connections themselves are in theory immune to this error, Runnables often use the Ensembl connection mechanism via various Ensembl APIs and might still get into trouble. The Core API especially has two evil parameters:
disconnect_when_inactive(boolean). When set the API will disconnect after every single query. This can result in exhausting the pool of ports available on the worker’s machine, leading to all processes on this machine failing to open a network connection. You can spot this when MySQL fails with the error code 99 (Cannot assign requested address). Leave this one to zero unless you use other mechanisms such asprevent_disconnectto prevent this from happening (see below).reconnect_when_lost(boolean). When set the API will ping the server before every query, which is expensive.
There are however some useful methods and tools in the Core API:
disconnect_if_idle. This method will ask the DBConnection object to disconnect from the database if possible (i.e. if the connection is not used for anything). Simple, but it does the job. Use this when you’re done with a database.prevent_disconnect. This method will run a piece of code withdisconnect_when_inactiveunset. Together they can form a clean way of handling database connections:Set
disconnect_when_inactivegenerally to 1 – This works as long as the database is used for just one query once in a while.Use
prevent_disconnectblocks when you’re going to use the database for multiple, consecutive, queries.
This way, the connection is only open when it is needed, and closed the rest of the time.
ProxyDBConnection. In Ensembl, a database connection is bound to one database only. However data can be spread across multiple databases on the same server (e.g. the Ensembl live MySQL server), and the API is going to create one connection for each database, potentially quickly exhausting the number of connections available, and the API is going to create one connection for each database, potentially quickly exhausting the number of connections available.ProxyDBConnectionis a way of pooling multiple database connections to the same server within the same object (connection). See an example in the Ensembl-compara API.